Structural Mass Spectrometry
Ion mobility-mass spectrometry (IM-MS) allows the separation of ionized molecules based on their structural properties such as size and shape, in addition to their mass-to-charge ratio. Combining ion mobility with mass spectrometry allows us to gain structural insight concerning our charged analyte species. During an ion mobility experiment, these charged species transverse a drift tube filled with a neutral buffer gas. The charged species experience elastic interactions with these neutral gas molecules. The larger the charged species the slower they transverses the drift tube. The time it takes for the charged species to transverse the drift tube can then be converted into a collision cross section value, which is representative of a rotationally averaged surface area.

A schematics of an ion mobility drift tube to show ions separating based on their size. The sample data shows that a shorter drift time corresponds to a smaller conformation, while a longer drift time corresponds to a larger conformation. Collision cross section values are then derived for these conformations providing a rotationally averaged surface area for the molecule.
A correlation can then be made between the mass-to-charge ratios and the collision cross section values resulting in a conformational landscape. Previous studies show that different classes of biomolecules separate out in this conformational landscape (oligonucleotides < carbohydrates < peptides < lipids) allowing for the separation and identification of these classes in complex biological samples, which are common in systems biology.1 These biological classes separate in conformational space due to their unique and differentiating structural characteristics.
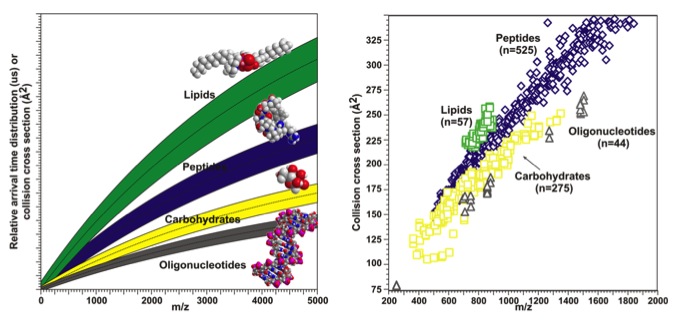
The data from an IM-MS experiment can be plotted as shown above generating a conformational space landscape. The mass-to-charge ratio is plotted against the CCS values. A cartoon of the data is shown on the left and actual data is plotted on the right. The different classes of biomolecules separate out in conformational space due to similar structural characteristics within their classes.
Structural IM-MS studies are often supplemented with computational work that samples the conformational space of the molecules of interests in silico. These computational studies consists of two steps: 1) Computationally sampling the conformational space of the molecule with molecular dynamics or other sampling methods, followed by 2) A theoretical determination of the CCS of each of these generated conformations. Experimental CCS values can then be used to discriminate the theoretically generated conformations. The resulting conformations can then be used to suggest possible structural characteristics that lead to the observed experimental CCS values.
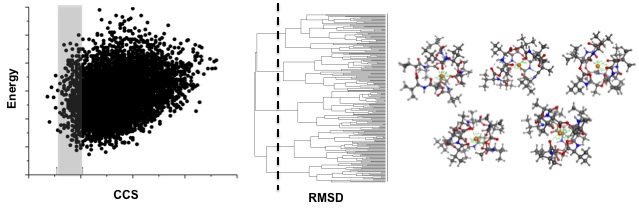
A sample computational workflow is shown to support IM-MS experimental measurements. Conformations are generated and then discriminated against using the experimentally derived CCS values. The resulting conformations can then be clustered based on RMSD and the resulting conformations can be examined to provide insight into why the molecules fall where they do in the conformational landscape.
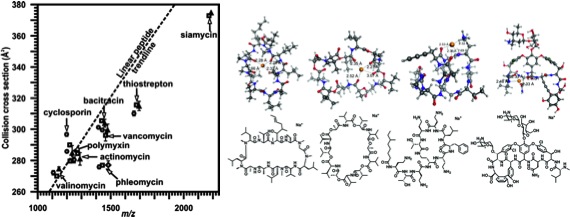
A study performed in our lab that used IM-MS and supporting computational methods to investigate why natural products deviate from linear peptide trendlines. 2
1. Fenn, L. S.; Kliman, M.; Mahsut, A.; Zhao, S. R.; McLean, J. A., Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples. Anal. Bioanal. Chem. 2009, 394 (1), 235-244.
2. Goodwin, C. R.; Fenn, L. S.; Derewacz, D. K.; Bachmann, B. O.; McLean, J. A., Structural Mass Spectrometry: Rapid Methods for Separation and Analysis of Peptide Natural Products. Journal of natural products 2012, 75 (1), 48-53.