Isomerization insights: a deeper understanding of Pin1-PPARγ dynamics
By Cameron I. Cohen
 Amino acid peptides can exist in cis and trans conformations, with most heavily favoring the latter. Proline, however, is unique among amino acids in that its peptide bonds can stably populate both conformations. The exchange, or isomerization, between cis and trans proline conformations is a relatively slow process which acts as a switch-like mechanism to modify protein activity. Enzymes known as peptidyl prolyl cis-trans isomerases (PPIases) catalyze the switch between cis and trans conformations to regulate processes such as the DNA damage response, gene transcription, and cellular localization.
Amino acid peptides can exist in cis and trans conformations, with most heavily favoring the latter. Proline, however, is unique among amino acids in that its peptide bonds can stably populate both conformations. The exchange, or isomerization, between cis and trans proline conformations is a relatively slow process which acts as a switch-like mechanism to modify protein activity. Enzymes known as peptidyl prolyl cis-trans isomerases (PPIases) catalyze the switch between cis and trans conformations to regulate processes such as the DNA damage response, gene transcription, and cellular localization.
Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (Pin1) is a multidomain PPIase with an N-terminal WW domain connected to a C-terminal PPIase domain by a flexible linker. The WW domain directs binding to protein targets containing a phosphorylated serine (pS) or threonine (pT) N-terminally adjacent to a proline, either of which increase the likelihood of that proline adopting a cis conformation. The PPIase domain is responsible for the isomerase activity. Target peptide binding to the WW domain has been shown to prime the PPIase domain for optimal catalytic activity, but a complete understanding of Pin1-target substrate interaction dynamics is lacking.
In particular, the structural basis for Pin1 binding to the substrate peroxisome proliferator-activated receptor gamma (PPARγ) is not well characterized. PPARγ is a lipid-sensing nuclear receptor which regulates genes associated with insulin resistance and the differentiation of mesenchymal cells into adipocytes. Studies have suggested Pin1 binds to either the AF-1 or LBD domain, but identification of the specific residues involved and the mechanism by which Pin1 accelerates cis-trans isomerization is needed to fully understand the regulation of PPARγ-mediated transcription.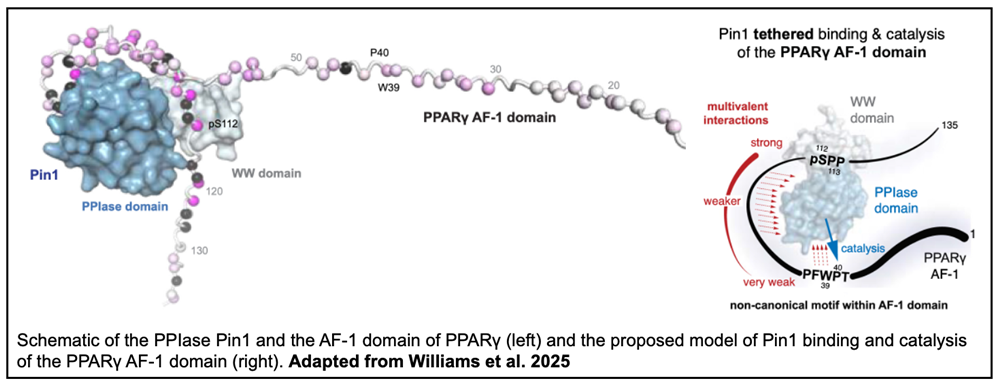
In this paper from the Kojetin laboratory, graduate student Christopher Williams, staff scientist Paola Munoz-Tello and a collaborator from the Scripps Institute use NMR spectroscopy to investigate Pin1- PPARγ binding. As both the AF-1 and LBD domains of PPARγ have been implicated in Pin1 binding, the researchers first sought to investigate the phosphorylation of pS-P motifs within the two domains. In-vitro phosphorylation followed by phos-tag SDS-PAGE revealed no phosphorylation of the LBD, but the technique did identify two distinct phosphorylation sites in AF-1. One site, S112, had been previously identified, but the possibility of a phosphorylated T75 site was a novel find.
However, when mass spectroscopy was performed on full-length PPARγ overexpressed in HEK293T cells, phosphorylation of T75 was not detected. While T75 may be phosphorylated in other cell lines or under different cellular conditions, the authors chose to focus solely on S112 for the remainder of the paper. Subsequently, the P76A PPARγ mutant was designed to eliminate confounding effects of the T75 site. NMR analysis was then conducted between Pin1 and PPARγ AF-1 in phosphorylated and unphosphorylated states. Strong chemical shift perturbations (CSPs) under the phosphorylated condition indicated that AF-1 phosphorylation enhances binding to Pin1.
To map the specific residues involved, the investigators analyzed NMR spectra of binding between either the WW and PPIase domains of Pin1 and phosphorylated/unphosphorylated AF-1. Notable CSPs demonstrated the WW domain of Pin1 specifically binds the S112 motif when AF-1 is phosphorylated and a tight binding affinity was fit to the titration data. The binding affinity between the WW domain and the unphosphorylated AF-1 could not be reliably calculated, likely due to weak binding. The PPIase domain of Pin1 also generated CSPs upon binding to AF-1, including around a PFWP motif previously shown to form interdomain contacts with the PPARγ LBD.
Additionally, binding of Pin1 to AF-1 accelerates the cis-trans isomerization of the WP dipeptide. Taken together, these data indicate that cooperative, allosteric mechanisms involving multiple surfaces underlie the interaction between Pin1 and PPARγ. Finally, the use of a pharmacological Pin1 inhibitor on HEK293T cells decreased PPARγ transcription to similar levels as a PFWP mutant, suggesting cis-trans isomerization of the WP dipeptide plays a key role in the regulation of PPARγ transcription. Altogether, this work contributes to a greater understanding of Pin1-mediated enzyme catalysis and has exciting implications for the understanding of PPARγ regulation.
To learn even more about Pin1-PPARγ dynamics check out the full paper in PNAS!
Leave a Response
You must be logged in to post a comment