Redefining synaptic architecture: nanoblocks as key organizing units in the postsynaptic density
 Cellular function relies on the nanoscale organization of molecular components. Pre-synaptic, post-synaptic and cell adhesion proteins within the synapse are carefully organized in nanostructures to facilitate the transmission of information between neurons, a process which is essential for synaptic plasticity, learning and memory formation. However, the precise structural arrangement of synapses is largely unknown due to their molecular complexity.
Cellular function relies on the nanoscale organization of molecular components. Pre-synaptic, post-synaptic and cell adhesion proteins within the synapse are carefully organized in nanostructures to facilitate the transmission of information between neurons, a process which is essential for synaptic plasticity, learning and memory formation. However, the precise structural arrangement of synapses is largely unknown due to their molecular complexity.
In this paper, postdoctoral fellow, Dr. Rong Sun, and members of the QJ Zhou lab use cryo-electron tomography (cryo-ET) to tackle this problem in the synapses of excitatory primary hippocampal neurons.
Cryo-electron tomography (cryo-ET) is a powerful technique which produces 3D volume reconstructions of samples and is capable of investigating biological structures in their near-native environment. In this project, the reconstructed tomograms revealed densities within the postsynaptic density (PSD), an electron-dense lamina just beneath the postsynaptic membrane. Analysis of the tomograms revealed five distinct density clusters within the PSD. To investigate these structures at an even higher resolution, the authors developed a process to isolate the synaptic terminals themselves from brain tissue.
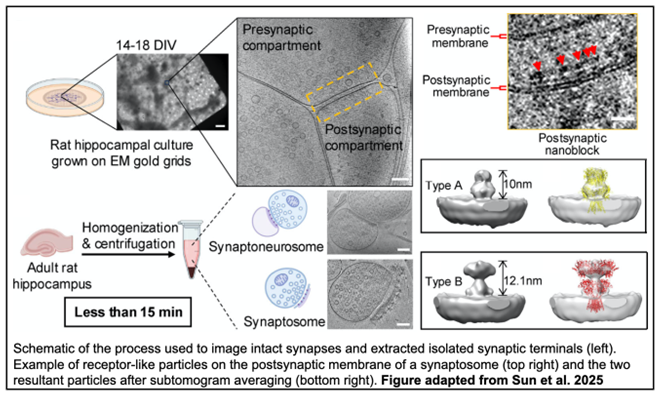 The preparations resulted in two types of isolated terminals: synaptoneurosomes, which retain an enclosed postsynaptic compartment, and synaptosomes, which retained a patch of postsynaptic membrane with PSD. In synaptosomes, the PSD also contained distinct density clusters, although synaptosomes had seven as opposed to the five seen in intact synapses. Additionally, synaptic vesicles in intact synapses were slightly larger than those in synaptosomes and synaptoneurosomes. However, all the synaptic vesicles fell within the average range of the 40-50 nm observed in excitatory neurons, indicating that isolated synaptic terminals maintain consistent structural characteristics. Interestingly, synaptosome clefts tend to be wider than synaptoneurosomes and intact synapses, suggesting that synaptosomes and synaptoneurosomes represent two stable states after isolation.
The preparations resulted in two types of isolated terminals: synaptoneurosomes, which retain an enclosed postsynaptic compartment, and synaptosomes, which retained a patch of postsynaptic membrane with PSD. In synaptosomes, the PSD also contained distinct density clusters, although synaptosomes had seven as opposed to the five seen in intact synapses. Additionally, synaptic vesicles in intact synapses were slightly larger than those in synaptosomes and synaptoneurosomes. However, all the synaptic vesicles fell within the average range of the 40-50 nm observed in excitatory neurons, indicating that isolated synaptic terminals maintain consistent structural characteristics. Interestingly, synaptosome clefts tend to be wider than synaptoneurosomes and intact synapses, suggesting that synaptosomes and synaptoneurosomes represent two stable states after isolation.
Having validated isolated terminals as a suitable model for studying synaptic organization, the authors then applied DBSCAN, a density-based clustering nonparametric algorithm, to further assess the PSD densities. Distinct clusters within the PSD were designated as “nanoblocks,” and the area and width of each nanoblock was determined. The size and distribution of the nanoblocks varied widely, but the patterns were consistent across synaptosomes, synaptoneurosomes, and intact synapses, indicating that these structures likely reflect synaptic architecture in vivo. The researchers then focused on ascertaining the composition of the nanoblocks, employing subtomogram averaging to detect postsynaptic receptor-like particles in synaptosomes. Two average structures were determined: a type A, which most closely resembled O-shaped AMPARs, and a type B, which most closely resembled Y-shaped AMPARs. Further, when analyzing the spatial distribution of particles, type A particles were found to be closer to nanoblocks than type B particles. Finally, the authors analyzed the distribution of nanoblocks relative to potential release sites, as determined by the presence of docked, tethered, or partially fused vesicles. While the PSD nanoblocks as a whole did not align with potential release sites, nanoblocks containing type A/B particles did, suggesting only certain nanoblocks participate in transsynaptic alignment.
Overall, this work has broadened our understanding of the nanoscale organization of neurons, specifically in the PSD of excitatory synapses. The discovery of heterogenous nanoblocks prompts questions about the relationship between spatial distribution, composition, and function, paving the way for exciting future research.
Check out the full story in the Journal of Cell Biology!
~ Cameron I.Cohen
Leave a Response
You must be logged in to post a comment