Liu, Qi; Wang, Yu; Hsu, Chihyuan; Wanjalla, Celestine N.; Lau, Ken S.; & Shyr, Yu. (2026). Microenvironment-aware spatial modeling for accurate inference of cell identity. Nucleic Acids Research, 54(1). https://doi.org/10.1093/nar/gkaf1477
Spatial omics technologies make it possible to measure many molecular features in cells while also preserving information about where those cells are located in a tissue. This spatial context provides valuable insight into how cells are organized and how tissues are structured. New platforms that work at single-cell resolution have further improved our ability to detect cell states that depend on the surrounding microenvironment. However, most existing computational tools for analyzing spatial omics data focus on identifying broad spatial regions rather than determining the identities of individual cells.
Traditional single-cell clustering methods define cell identities using only molecular features inside the cell, such as gene expression, and do not account for how nearby cells and the local tissue environment influence cell behavior. To address this limitation, we introduce MEcell, a method that directly incorporates spatial information and automatically determines how much influence the surrounding environment should have when identifying cell types. MEcell does not require users to tune parameters, making it easier to apply across datasets.
We tested MEcell on 90 simulated datasets and 7 real-world datasets from multiple spatial transcriptomics platforms and tissue types, including MERFISH Vizgen, Xenium, CosMx, Visium HD, Slide seq V2, and open ST. Across all tests, MEcell consistently performed better than existing methods at accurately identifying cell identities. These results show that the local microenvironment plays a crucial role in defining cell identity and demonstrate that MEcell is a powerful tool for capturing the full diversity of cells in spatial omics data.
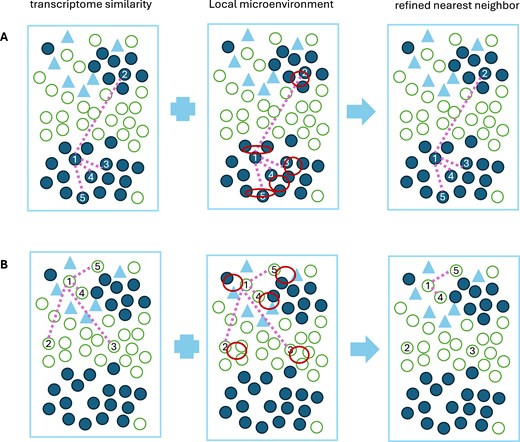
Figure 1.
The rationale of MEcell. (A) A toy example where a cell’s transcriptionally similar neighbors are located within the same microenvironment, indicating that the microenvironment will play a minimal role in shaping the nearest-neighbor graph. (B) A toy example where a cell’s transcriptionally similar neighbors are located across distinct microenvironments, suggesting that the microenvironment will play a significant influence in shaping the nearest-neighbor graph.