Clicking into place with paramagnetic NMR restraints

Jens Meiler, Distinguished Research Professor
Nuclear magnetic resonance (NMR) is an incredibly powerful technique for determining protein structure. A complete dataset of chemical shifts (CS) and nuclear Overhauser effects (NOEs) obtained from NMR can be used to determine structural models of a protein of interest.
In the case of integral membrane proteins, however, there is a lot of difficulty in obtaining complete datasets. NOEs can only be measured at proton-proton distances of ~5 Å but transmembrane helix-helix distances are typically in the range of 10-11 Å. To circumvent this problem, paramagnetic tags have been developed, which, when added to the target protein, produce NMR observables such as pseudocontact shifts (PCSs). The conventional method of introducing paramagnetic tags relies on disulfide linkages with site-specific cysteine residues, but this requires the prior removal of all native cysteine residues. As many proteins rely on disulfide bonds for structural stability and function, this process is not widely applicable.
In a recent study published in the Journal of Biomolecular NMR, Kaitlyn Ledwitch, research assistant professor in the Meiler lab, and colleagues detail the development of a novel paramagnetic spin-label system which relies on the use of non-canonical amino acids in the protein of interest. Following site-specific incorporation of p-azido-ʟ-phenylalanine residues, C3-lanthanide tags are attached with a copper-catalyzed click chemistry reaction. Data acquired from C3-tagged residues can be used to produce a sparse PCS database which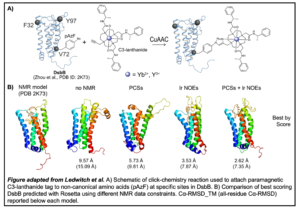 can be used as constraints in Rosetta protein structure predictions. When PCS data are combined with long-range NOEs, model accuracy, sampling and scoring is greatly improved.
can be used as constraints in Rosetta protein structure predictions. When PCS data are combined with long-range NOEs, model accuracy, sampling and scoring is greatly improved.
This system allows for the collection of detailed NMR data without the need to disrupt native cysteines and disulfide bonds, which could provide insight into the structural details of previously unexplored proteins.
Read more about the study here! ~Cameron I. Cohen
Leave a Response
You must be logged in to post a comment