Research Spotlight: Egli Lab
Martin Egli, Biochemistry, and Joel Harp, Crystallography Facility Director, have combined X-ray and neutron crystallography to investigate the mechanics of how hydration affects DNA structure. This joint approach is important as structures determined using X-rays can only resolve the water molecules as spheres of electron density around the oxygen atom. Neutrons are scattered by atomic nuclei, which allows for the determination of the orientation of water molecules, especially if deuterated water is used for better resolution.
Here is the science behind the approach…
Due to its highly ionic character, the structure of DNA is strongly dependent on hydration. Even high-resolution X-ray crystal structures provide information on the location of waters only as a sphere of electron density around the oxygen atom with little evidence of the presence of the two hydrogens. X-rays are scattered by electrons and nearly blind to the presence of hydrogen with its single, loosely bound electron in a water molecule. Neutrons on the other hand are scattered by atomic nuclei. The scattering cross-section, scattering power, of an atom in a neutron beam is complex and counter-intuitive. In particular, hydrogen has a negative scattering cross-section and exceptionally high incoherent scattering that contributes background in a neutron diffraction image. The hydrogen isotope, 2H or deuterium, has a positive scattering cross-section in the same range as oxygen and carbon and very little incoherent scattering. Using deuterated water, D2O, we are able to not only see the location of water molecules but also to experimentally determine their orientation and examine hydrogen bonding networks to explain the mechanics of how hydration affects DNA structure. We focused on the left-handed Z-DNA duplex [d(CGCGCG)]2 because it exhibits intriguing inter-base and base-phosphate hydration patterns in the minor groove and on the convex surface. Z-DNA crystals diffract to high resolution and are easily grown to the large size necessary for neutron diffraction experiments.
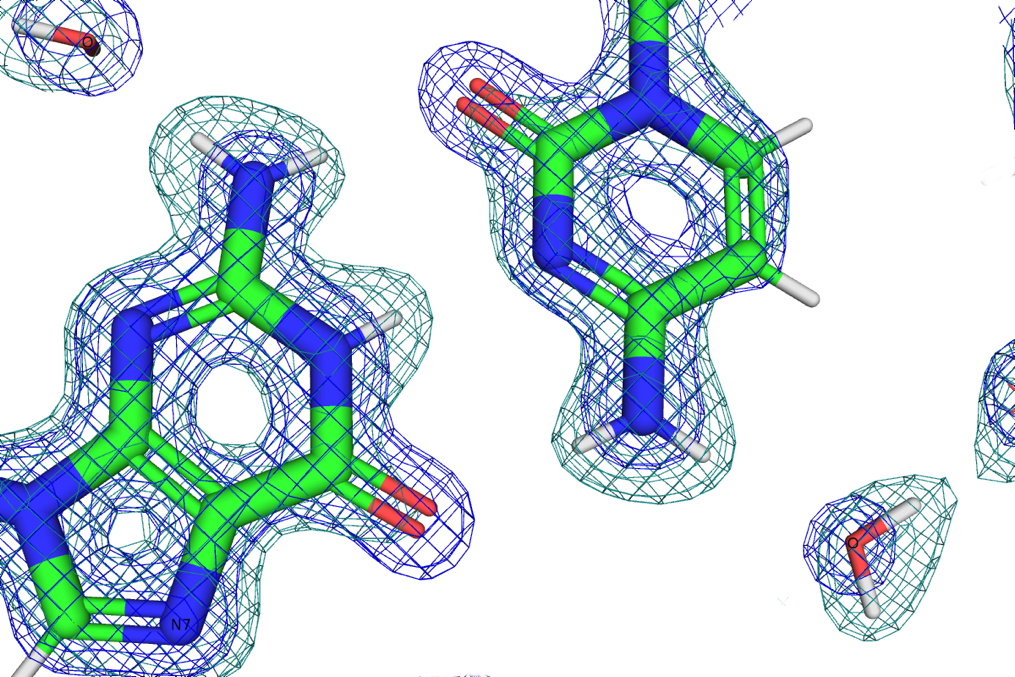
To amplify the value of X-ray and neutron crystallography, we refined the structure using data sets from our in-house MetalJet X-ray source and the MaNDi neutron beamline in a joint X-ray/neutron refinement to get a more complete picture. The Macromolecular Neutron Diffraction beamline at the Spallation Neutron Source (MaNDi/SNS), located a short and scenic drive away from Vanderbilt University at Oak Ridge National Laboratory (ORNL), provides the world’s most intense source of neutrons as well as an excellent sample environment for low temperature, cryogenic, data collection. Cryogenic data collection is essential for slowing molecular motion to provide better data and germane to studying the orientations of first- and second-shell water molecules.
Leave a Response
You must be logged in to post a comment