Zhu, Junchao; Deng, Ruining; Guo, Junlin; Yao, Tianyuan; Xiong, Juming; Qu, Chongyu; Yin, Mengmeng; Wang, Yu; Zhao, Shilin; Yang, Haichun; Xu, Daguang; Tang, Yucheng; & Huo, Yuankai. (2025). Img2ST-Net: Efficient high-resolution spatial omics prediction from whole-slide histology images via fully convolutional image-to-image learning. Journal of Medical Imaging, 12(6), 61410. https://doi.org/10.1117/1.JMI.12.6.061410
Recent progress in multimodal artificial intelligence has shown that spatial transcriptomics data, which measure where genes are active within tissue, can potentially be generated from standard histology images, reducing the cost and time required for specialized experiments. However, newer spatial transcriptomics platforms such as Visium HD operate at very high resolution, down to about 8 micrometers, which creates major computational challenges. At this scale, traditional methods that predict gene expression one spot at a time become slow, unstable, and poorly suited to the extreme sparsity of gene expression, where many genes have very low or zero signal. To address this, the authors developed Img2ST Net, a high-resolution framework that predicts spatial transcriptomics data from histology images using a fully convolutional neural network, meaning the model generates dense gene expression maps all at once rather than sequentially. The method represents high-resolution spatial transcriptomics as groups of small regions called super pixels and reframes the task as an image generation problem with hundreds or thousands of output channels, each corresponding to a gene. This approach improves efficiency and better preserves spatial structure in the tissue. To evaluate performance under sparse expression conditions, the authors also introduced SSIM ST, a structural similarity-based metric designed specifically for high-resolution spatial transcriptomics. Testing on public breast and colorectal cancer Visium HD datasets at 8 and 16 micrometer resolution showed that Img2ST Net outperformed existing methods in both prediction accuracy and spatial coherence, while reducing training time by up to 28 times compared with spot-based approaches. Additional analyses showed that contrastive learning further improved spatial fidelity. Overall, this work provides a scalable and biologically meaningful solution for predicting high-resolution spatial transcriptomics data and supports future large-scale and resolution-aware spatial omics modeling.
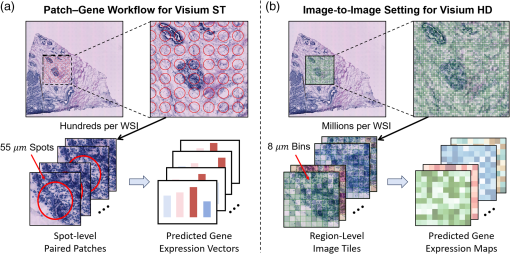
Fig. 1
Modeling paradigm for ST prediction. (a) Conventional patch-to-spot regression manner for Visium ST data: each WSI contains hundreds of 55μm spots for the ST slide. A separate gene expression vector is predicted for each spot from its corresponding image patch. (b) Our proposed image-to-image prediction framework for Visium HD data: each WSI contains millions of 8μm bins for the HD slide. A region-wise modeling strategy where each image region covers multiple bins is used to predict a high-resolution gene expression map, which enables more fine-grained and computationally efficient inference.