Newlin, Nancy R.; Kim, Michael E.; Kanakaraj, Praitayini; McMaster, Elyssa M.; Cho, Chloe; Gao, Chenyu; Hohman, Timothy J.; Beason-Held, Lori L.; Resnick, Susan M.; O’Bryant, Sid E.; Phillips, Nicole R.; Barber, Robert Clinton; Bennett, David Alan; Barnes, Lisa Laverne; Biber, Sarah A.; Johnson, Sterling C.; Archer, Derek B.; Li, Zhiyuan; Zuo, Lianrui; Moyer, Daniel C.; & Landman, Bennett Allan. (2025). Harmonizing 10,000 connectomes: Site-invariant representation learning for multi-site analysis of network connectivity and cognitive impairment. Journal of Medical Imaging, 12(6), 64001. https://doi.org/10.1117/1.JMI.12.6.064001
Diffusion magnetic resonance imaging data collected across different studies often vary because of differences in scanners, software, and acquisition protocols, which can introduce unwanted technical effects that interfere with biological analysis. This is especially challenging in large multi site studies of Alzheimer’s disease, where both technical variation and true disease related changes are present. In this study, the authors developed a harmonization approach that learns low dimensional representations of brain structural connectivity that are insensitive to imaging site, scanner type, and acquisition settings, while still preserving information related to cognitive status. They used a conditional variational autoencoder, a type of deep learning model that compresses data into a latent space while controlling which information is retained or removed. The model was trained on diffusion imaging data from 6956 individuals across 9 cohorts and 38 imaging sites, totaling nearly 12,000 scans, including participants with normal cognition, mild cognitive impairment, and Alzheimer’s disease. The learned representations successfully removed statistically significant site related effects across 12 brain network connectivity measures and improved the accuracy of predicting cognitive diagnosis from 68 percent to 73 percent. The method showed consistent performance across multiple data configurations, demonstrating that representation learning can effectively reduce scanner related confounding while strengthening biologically meaningful signals in large neuroimaging studies.
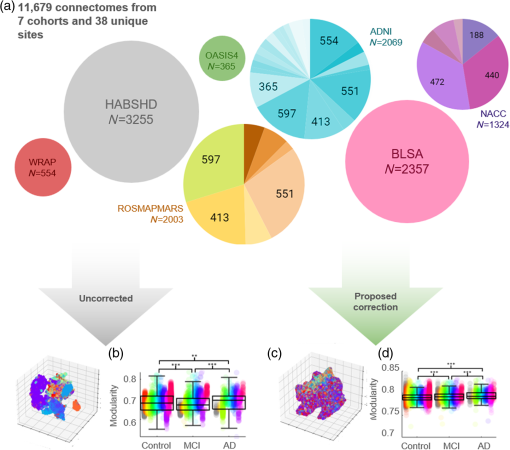
Fig. 1
A total of 11,927 connectomes are pooled together across 9 cohorts and 35 unique imaging acquisitions (for a total of 38 “sites”). (a) If we naively combine all data and create t-SNE plots, there are obvious clusters driven by site. We compare cognitive diagnosis differences in one network connectivity measure, brain modularity, computed from these uncorrected connectomes. (b) Site-wise differences drive the detected differences and result in false findings. (c) In comparison, the t-SNE features of our learned representation have decreased site information, and the (d) resulting modularity values are harmonized and provide clear, significant (p<0.001) trends with cognitive diagnosis.