Guo, J., Lu, S., Cui, C., Deng, R., Yao, T., Tao, Z., Lin, Y., Lionts, M., Liu, Q., Xiong, J., Wang, Y., Zhao, S., Chang, C. E., Wilkes, M., Fogo, A. B., Yin, M., Yang, H., & Huo, Y. (2025). Evaluating cell AI foundation models in kidney pathology with human-in-the-loop enrichment. Communications Medicine, 5(1), 495. https://doi.org/10.1038/s43856-025-01205-x
Large artificial intelligence foundation models are becoming important tools in healthcare, including digital pathology, where they help analyze medical images. Many of these models have been trained to handle complex tasks such as diagnosing diseases or measuring tissue features using very large and diverse datasets. However, it is less clear how well they perform on more focused tasks, such as identifying and outlining cell nuclei within images from a single organ like the kidney. This study examines how well current cell foundation models perform on this task and explores practical ways to improve them.
To do this, the researchers assembled a large dataset of 2,542 kidney whole slide images collected from multiple medical centers, covering different kidney diseases and even different species. They evaluated three widely used, state-of-the-art cell foundation models—Cellpose, StarDist, and CellViT—for their ability to segment cell nuclei. To improve performance without requiring extensive, time-consuming pixel-level annotations from experts, the team introduced a “human-in-the-loop” approach. This method combines predictions from multiple models to create higher-quality training labels and then refines a subset of difficult cases with corrections from pathologists. The models were fine-tuned using this enriched dataset, and their segmentation accuracy was carefully measured.
The results show that accurately segmenting cell nuclei in kidney pathology remains challenging and benefits from models that are more specifically tailored to this organ. Among the three models, CellViT showed the best initial performance, with an F1 score of 0.78. After fine-tuning with the improved training data, all models performed better, with StarDist reaching the highest F1 score of 0.82. Importantly, combining automatically generated labels from foundation models with a smaller set of pathologist-corrected “hard” image regions consistently improved performance across all models.
Overall, this study provides a clear benchmark for evaluating and improving cell AI foundation models in real-world pathology settings. It also demonstrates that high-quality nuclei segmentation can be achieved with much less expert annotation, supporting more efficient and scalable workflows in clinical pathology without sacrificing accuracy.
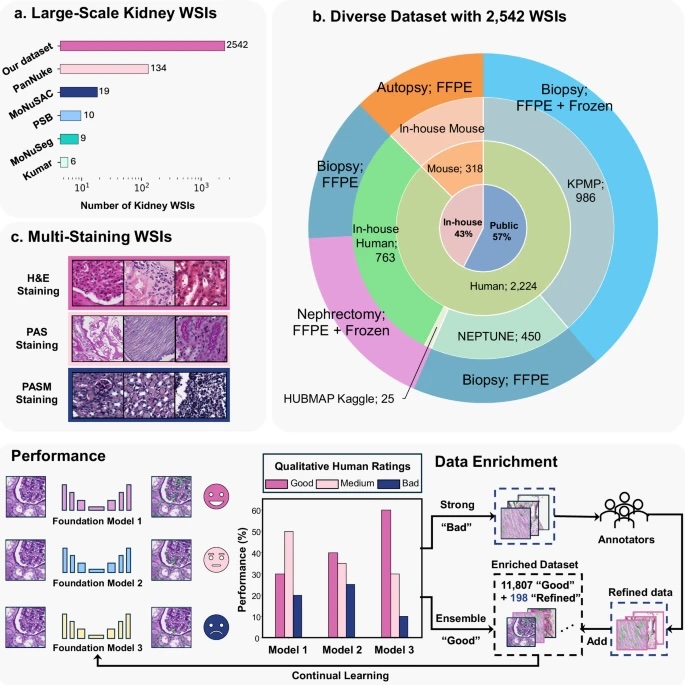
Fig. 1: Overall framework.
The upper panel (a–c) illustrates the diverse evaluation dataset consisting of 2542 kidney WSIs. a shows the number of kidney WSIs in publicly available cell nuclei datasets versus our evaluation dataset, which exceeds existing datasets by a large margin. b depicts the diverse data sources included in our dataset. c indicates that these WSIs were stained using Hematoxylin and Eosin (H&E), Periodic acid–Schiff methenamine (PASM), and Periodic acid–Schiff (PAS). Performance: Kidney cell nuclei instance segmentation was performed using three SOTA cell foundation models: Cellpose, StarDist, and CellViT. Model performance was evaluated based on qualitative human feedback for each prediction mask. Data Enrichment: A human-in-the-loop (HITL) design integrates prediction masks from performance evaluation into the model’s continual learning process, reducing reliance on pixel-level human annotation.