Krishnan, Aravind R., Li, Thomas Z., Remedios, Lucas W., Kim, Michael E., Gao, Chenyu., Rudravaram, Gaurav., McMaster, Elyssa M., Saunders, Adam M., Bao, Shunxing., Xu, Kaiwen., Zuo, Lianrui., Sandler, Kim Lori., Maldonado, Fabien., Huo, Yuankai., & Landman, Bennett Allan. (2025). Multipath cycleGAN for harmonization of paired and unpaired low-dose lung computed tomography reconstruction kernels. Medical Physics, 52(11), e70120. https://doi.org/10.1002/mp.70120
CT scans can look noticeably different depending on the reconstruction kernel used to process the images. These kernels change how sharp or noisy an image appears, which can lead to big differences in important measurements—such as how much emphysema is present in the lungs. While it’s fairly easy to make images consistent when they come from the same type of scanner, this becomes much harder in studies that collect scans from many hospitals and manufacturers. Because each manufacturer uses different kernels, the measurements can become inconsistent, making it difficult to compare results. To fix this, we need a way to standardize all CT images so they look as if they were created using the same reference kernel.
In this study, we tested whether we could train a computer model to do this standardization using both paired data (scans from the same person processed with two different kernels from one manufacturer) and unpaired data (scans from different people and different manufacturers). Our goal was to use both types of data to create a shared representation of the images that allows for consistent comparisons across all scanners.
We created a deep learning model called a multipath cycleGAN, which can learn how to “translate” CT images from one kernel style to another. It uses a shared latent space (a common internal representation), along with several encoder–decoder pathways and discriminators that help the model learn from both paired and unpaired examples. We trained the model using CT scans from seven common reconstruction kernels from the National Lung Screening Trial, giving us 42 possible kernel combinations to harmonize.
We then tested the model using hundreds of additional scans. For paired kernels, we looked at whether the model reduced differences in percent emphysema, and it did—performing better than comparison methods in several cases. For unpaired kernels, we converted all scans to look like they were processed with a reference soft kernel or a reference hard kernel and again measured emphysema levels. Our model reduced differences in many kernel types and performed similarly or better than existing approaches. We also checked whether harmonization preserved important anatomical structures such as lung vessels, muscle, and fat, and found that our method generally maintained these details.
Overall, our results show that combining paired and unpaired data in a shared latent space multipath cycleGAN can reduce errors in emphysema measurement and keep anatomical structures consistent. This approach offers a promising way to make CT scans from different scanners and reconstruction kernels more comparable, which is important for large research studies and long-term patient monitoring.
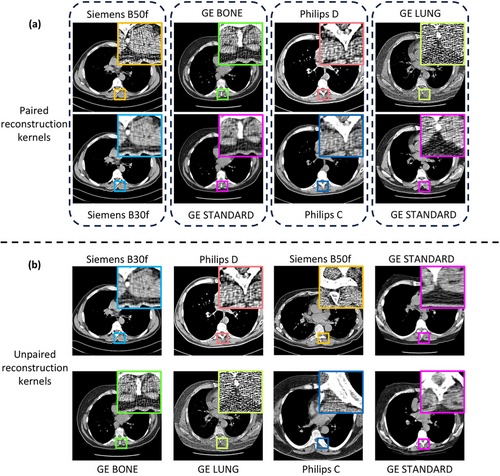
FIGURE 1
Reconstruction kernels influence the noise and resolution of the underlying anatomical structure in a computed tomography image. (a) Paired reconstruction kernels obtained from a given vendor exhibit a one-to-one pixel correspondence between the scans, which enables kernel harmonization. However, (b) across vendors, unpaired kernels show differences in anatomy, scan protocol, field of view, and reconstruction window. This creates additional difficulties that make harmonization a more challenging task.