Hu, Yunfei., Lin, Zhenhan., Xie, Manfei., Yuan, Weiman., Li, Yikang., Rao, Mingxing., Liu, Yichen Henry., Shen, Wenjun., Zhang, Lu., & Zhou, Xin Maizie. (2025). MaskGraphene: an advanced framework for interpretable joint representation for multi-slice, multi-condition spatial transcriptomics. Genome Biology, 26(1), 380. https://doi.org/10.1186/s13059-025-03850-w
Recent advances in spatial transcriptomics (ST), which maps gene activity across tissue, show the need to analyze multiple tissue slices together. A major challenge is creating meaningful representations (embeddings) that preserve the tissue’s spatial layout while correcting for differences between slices (batch effects). We introduce MaskGraphene, a graph neural network that integrates ST data using masked self-supervised learning, triplet loss, and cluster-wise local alignment. By forming indirect “soft-links” and direct “hard-links” between slices, MaskGraphene produces joint embeddings that closely preserve spatial geometry. Compared to eight existing methods, MaskGraphene achieves better alignment and interpretability. It also improves downstream analyses, including identifying distinct tissue domains, reconstructing developmental trajectories, discovering biomarkers, and mapping brain layers, providing a robust tool for integrating ST data and extracting biological insights.
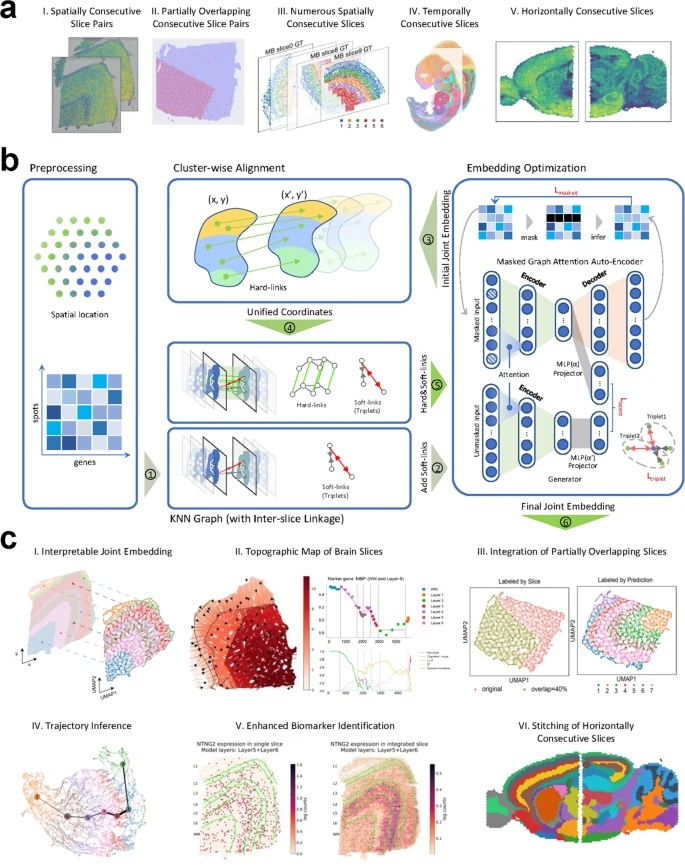
Fig 1
MaskGraphene workflow. a Illustration of spatial transcriptomics data integration scenarios addressed by MaskGraphene, including spatially consecutive slice pairs (I), simulated partially overlapping consecutive slice pairs (II), numerous spatially consecutive slices (III), temporally consecutive slices (IV), and horizontally consecutive slices (V). b Workflow of MaskGraphene: The preprocessing step organizes spatial coordinates and gene expression data. Inter-slice linkage is established through the construction of “hard-links” via cluster-wise local alignment and “soft-links” using contrastive learning with triplets. Embedding optimization leverages a masked graph autoencoder to generate batch-corrected joint embeddings by optimizing masked self-supervised loss () and triplet loss. Steps 1-5 of the main method are indicated with numbered circles. c Applications and evaluations (Step 6): (I) Interpretable joint embedding captures the original geometric structure. (II) Topographic map of brain slices with isodepth analysis reveals gene expression gradients across cortical layers. (III) Validation with simulated data demonstrates robust integration of partially overlapping slices. (IV) Trajectory inference reveals linearly connected developmental trends. (V) Alignment and integration of embryonic tissue structures enhance biomarker identification. (VI) Stitching of horizontally consecutive slices reconstructs spatially coherent regions