Wang, Yu, Yang, Haichun, Deng, Ruining, Huo, Yuankai, Liu, Qi, Shyr, Yu, & Zhao, Shilin. (2025). StImage: A versatile framework for optimizing spatial transcriptomic analysis through customizable deep histology and location informed integration. Briefings in Bioinformatics, 26(5), bbaf429. https://doi.org/10.1093/bib/bbaf429
Spatial transcriptomics (ST) is a technique that links gene activity with the physical location of cells in tissue, providing detailed insights into how tissues function. Current methods either focus on cell location or tissue images, but none fully combine gene expression, histology (tissue structure), and precise spatial information in a single framework. These methods also often perform inconsistently across different datasets. To address this, we developed stImage, an open-source R package that offers a flexible and comprehensive approach to ST analysis. stImage uses deep learning to extract features from tissue images and provides 54 strategies to integrate gene expression, tissue structure, and spatial data. We show that stImage works effectively across multiple datasets and helps users choose the best integration approach using a diagnostic graph. Overall, stImage improves the analysis of spatial transcriptomics data, enhancing our understanding of tissue organization. It is freely available at https://github.com/YuWang-VUMC/stImage.
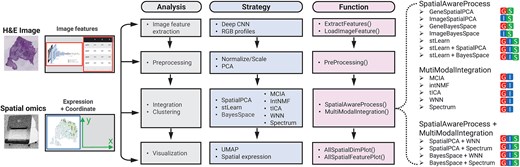
Figure 1
Workflow of stImage. stImage comprises four main steps, image features extraction, preprocessing, data integration, and visualization. The different color squares with abbreviation of different modalities show what combination of modalities were processed for that strategy: G for gene expression; S for spatial coordinates; I for image features.