Kuang, Xiaohan; Liu, Yunchao Lance; Lin, Xiaobo; Spencer-Smith, Jesse; Derr, Tyler; Wu, Yinghao; Bitter, Hans; Hu, Yongbo; Meiler, Jens; & Su, Zhaoqian. (2025). Superwater as a generative AI framework to predict water molecule positions on protein structures. Communications Chemistry, 8(1), 397. https://doi.org/10.1038/s42004-025-01789-4
Water molecules are essential for keeping proteins structurally stable and for enabling proteins to interact with other molecules. Accurately predicting where water molecules are located around a protein is important for understanding protein function and has major implications for protein engineering and drug discovery. This study introduces SuperWater, a new generative artificial intelligence framework designed to predict water molecule positions around protein structures with high accuracy. SuperWater combines a score-based diffusion model, which learns how molecular structures are formed and refined, with equivariant graph neural networks, which are neural networks designed to respect the three-dimensional shape and physical symmetry of proteins. By using both approaches together, SuperWater can more precisely model how water molecules arrange themselves around proteins. The method outperforms existing approaches, achieving state-of-the-art results in crystal water coverage, meaning how well it recovers water molecules seen in experimental crystal structures, as well as prediction precision. On average, SuperWater predicts water positions within 0.3 plus or minus 0.06 angstroms of experimentally measured locations. The framework is demonstrated in examples involving protein hydration, protein ligand binding, and protein-protein interaction sites. Overall, SuperWater is a flexible tool that can be applied to many areas, including structural biology, binding site prediction, multi-body docking, and water-mediated drug design.
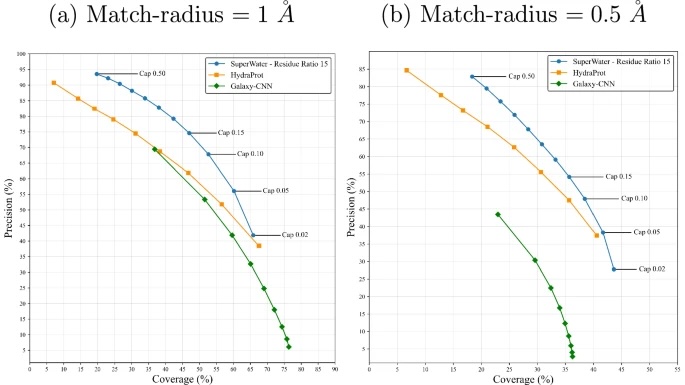
Fig. 1: Precision-coverage trade-off for the three methods.
Comparison of SuperWater (blue), HydraProt (orange), and GalaxyWater-CNN (green) in predicting water molecule positions at two matching radius: a 1.0 Å and b 0.5 Å. Precision is the fraction of predicted waters matching experimental waters within the given radius; coverage is the fraction of experimental waters matched by predictions. Curves are obtained by varying each model’s internal confidence probability threshold (cap).