Li, Qing; Song, Qingyuan; Chen, Zhishan; Choi, Jung-yoon; Moreno, Victor R.; Ping, Jie; Wen, Wanqing; Li, Chao; Shu, Xiang; Yan, Jun; Shu, Xiaoou; Cai, Qiuyin; Long, Jirong; Huyghe, Jeroen R.; Pai, Rish K.; Gruber, Stephen Bernard; Yang, Yaohua; Casey, Graham R.; Wang, Xusheng; Toriola, Adetunji T.; Li, Li; Singh, Bhuminder; Lau, Ken S.; Zhou, Li; Zhang, Zichen; Wu, Chong; Peters, Ulrike; Zheng, Wei; Long, Quan; Yin, Zhijun; & Guo, Xingyi. (2026). Large-scale integration of omics and electronic health records to identify potential risk protein biomarkers and therapeutic drugs for cancer prevention. American Journal of Human Genetics, 113(1), 41–56. https://doi.org/10.1016/j.ajhg.2025.11.008
Finding the right proteins to target and the drugs that act on them is essential for preventing cancer. In this study, we combined and closely analyzed data from large genome-wide association studies covering six common cancers: breast, colorectal, lung, ovarian, pancreatic, and prostate cancer. We identified 710 genetic variants that are independently linked to cancer risk. By connecting these variants to protein quantitative trait loci using blood-based proteomics data from more than 75,000 people, we found 365 proteins associated with cancer risk.
Further analysis showed that 101 of these proteins are very likely to play a direct role in cancer development, including 74 that have not been reported before. Among them, 36 proteins appear to be potentially druggable, meaning they could be targeted by existing or future medications. To explore real-world effects, we analyzed more than 3.5 million electronic health records and carried out emulated clinical trials comparing 11 commonly used drugs across 290 scenarios. We identified three drugs that were associated with a lower risk of colorectal cancer, including caffeine compared with paroxetine, haloperidol compared with prochlorperazine, and trazodone hydrochloride compared with paroxetine. In contrast, caffeine was linked to a higher cancer risk when compared with finasteride for colorectal cancer and fluoxetine for breast cancer.
A combined analysis across studies identified six drugs that were significantly associated with cancer risk. One of these, acetazolamide, was associated with a reduced risk of colorectal cancer. Overall, this study uncovers previously unknown protein biomarkers and potential drug targets across six major cancer types and highlights several already approved drugs that may have promise for cancer prevention.
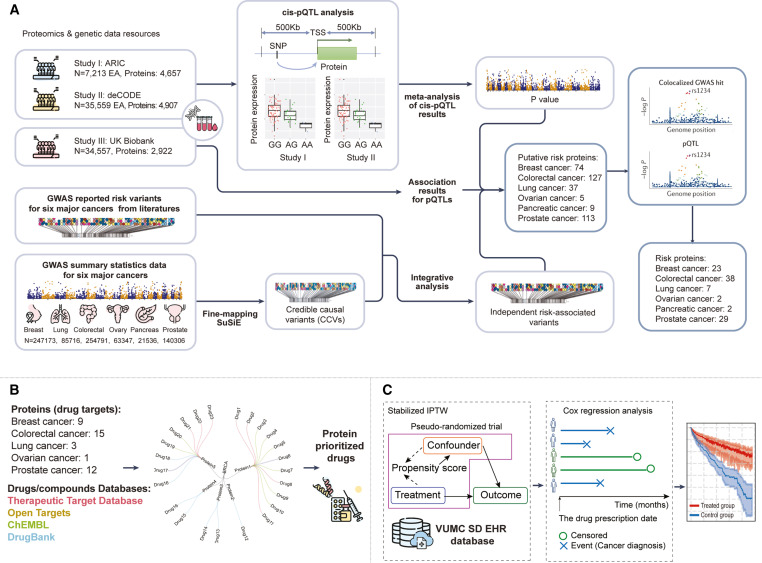
Figure 1 Overview of the analytical framework
(A) An illustration depicting the identification of proteins associated with the risk of the six major cancers: breast, lung, colorectal, ovarian, pancreatic, and prostate. Population-based proteomics data (for pQTLs) and GWAS data resources (for identifying lead variants) utilized in this study are shown on the left. Meta-analyses of cis-pQTLs from ARIC and deCODE, conducted through the SOMAscan platform, were combined with pQTL results from the UKB-PPP to identify potential risk proteins, as depicted in the middle images. Colocalization analyses between GWAS summary statistics and cis-pQTLs were performed to identify cancer risk proteins with high confidence, as illustrated on the right.
(B) The proteins with evidence of colocalization annotated based on drug-protein information from four databases: DrugBank, ChEMBL, TTD, and Open Targets.
(C) The framework for evaluating the effects of drugs approved for indications on cancer risk. The inverse probability of treatment weighting (IPTW) framework was utilized to construct emulations of treated-control drug trials based on millions of patients’ electronic health records stored at VUMC SD (left). In these emulations, the Cox proportional hazard model was conducted for each trial to assess the hazard ratio (HR) of cancer risk between the treated focal drug and the control drug (right).