Janve, Vaibhav A.; Seto, Mabel; Sperling, Reisa Anne; Aisen, Paul S.; Rissman, Robert A.; Koran, Mary Ellen Irene; Dumitrescu, Logan C.; Buckley, Rachel F.; & Hohman, Timothy J. (2025). Blood gene expression network expression strongly relates to brain amyloid burden. Alzheimer’s and Dementia, 21(12), e70982. https://doi.org/10.1002/alz.70982
Amyloid deposition, the buildup of amyloid beta protein in the brain, begins decades before clinical symptoms appear in Alzheimer disease. To understand early biological changes linked to amyloid, this study combined blood transcriptomics, which measure gene expression in whole blood, with positron emission tomography PET imaging that detects brain amyloid. The analysis included 1,739 cognitively unimpaired participants from the Anti Amyloid Treatment in Asymptomatic Alzheimer Disease A4 study. Whole blood RNA sequencing data were used to define groups of co expressed genes called gene modules, and linear regression models tested whether module expression was associated with amyloid PET burden while adjusting for age, sex, education, and apolipoprotein E APOE ε2 and ε4 genotypes.
Among 18 gene modules examined, one module enriched for histone genes located on chromosome 6 was significantly associated with amyloid burden, with higher amyloid levels linked to lower expression of this histone gene cluster. Histone genes encode proteins involved in DNA packaging and regulation of gene expression. In addition, estimated immune cell proportions derived from the blood transcriptomic data showed suggestive associations, including lower predicted levels of activated natural killer NK cells and CD4 positive activated memory T cells with higher amyloid deposition.
Overall, these findings identify the histone gene cluster on chromosome 6 and specific immune cell signatures as blood based correlates of brain amyloid accumulation in preclinical Alzheimer disease, suggesting that peripheral gene expression and immune changes may reflect early disease processes long before symptoms develop.
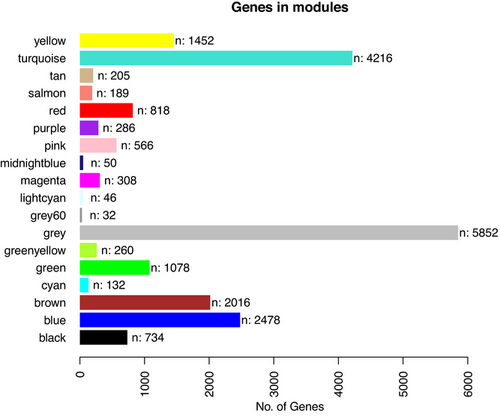
FIGURE 1
Gene expression modules determined with Weighted Gene Co-expression Network Analysis analysis. The gene module is presented along the y-axis. The number of genes is presented along the x-axis.