Semler, E. M., Michell, D. L., Kingsley, P. J., Ramirez, M. A., Massick, C., Castleberry, M., Doran, A. C., Carr, J. J., Marnett, L., Sheng, Q., Linton, M. F., & Vickers, K. C. (2026). Post-transcriptional modifications on tRNA fragments confer functional changes to high-density lipoproteins in atherosclerosis. Atherosclerosis, 412, 120584. https://doi.org/10.1016/j.atherosclerosis.2025.120584
Chemical modifications on RNA, called epitranscriptomic modifications, help control how RNA works in our cells. Small RNA fragments, known as tRNA-derived fragments (tDRs), can carry these modifications from their parent RNA. Some of these modified RNAs travel through the blood inside high-density lipoproteins (HDL), the same particles that carry “good cholesterol.” These HDL-associated RNAs (HDL-sRNAs) can influence immune responses and may play a role in atherosclerotic cardiovascular disease (ASCVD).
In this study, we compared HDL-sRNAs from healthy people and patients with advanced ASCVD. Using methods like LC-MS/MS, ARM-seq, and qPCR, we found that HDL from ASCVD patients had more RNA modifications. We identified a specific fragment, tDR-ArgACG, carrying a modification called 1-methyladenosine (m1A), which was more common in ASCVD. When these modified RNAs entered macrophages, immune cells that drive inflammation in arteries, they changed gene activity, including increasing TMEM123, a protein that helps immune cells move and stick to tissues. Experiments showed that HDL carrying m1A-tDR-ArgACG boosted TMEM123 in macrophages, and blocking the m1A modification reduced this effect.
These findings suggest that HDL can deliver modified RNAs to immune cells, triggering inflammation in atherosclerotic plaques. This reveals a new way that HDL-sRNAs may contribute to cardiovascular disease and points to potential targets for therapy.
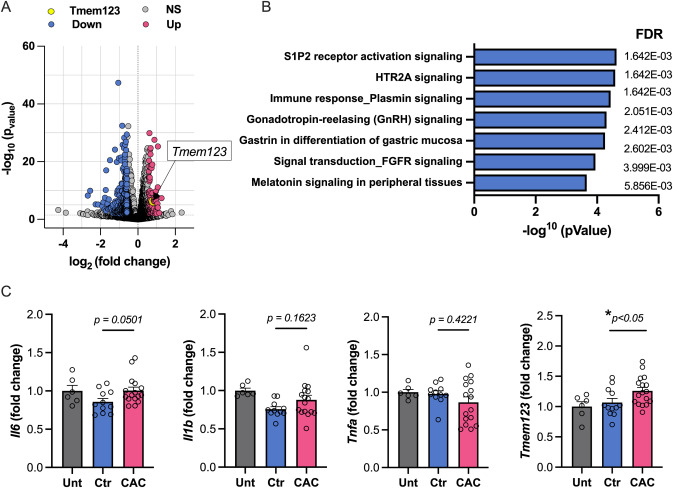
Fig. 1 CAC HDL induces non-classical activation and select immune response pathway and gene expression changes in macrophages (A) Volcano plot depicting log10 p-adjusted (Q)-value versus log2 fold changes for all differentially expressed (≥2.0 absolute fold change) genes in BMDM’s treated with CAC + HDL or Ctr-HDL (n = 3). (B) EGR1-containing gene set pathway analyses (Meta Core) enrichment, ranked by -log10(p-value) with false discovery rate (FDR) shown. (C) mRNA expression by qPCR displaying fold change from BMDMs treated with Ctr-HDL (n = 9) and CAC+HDL (n = 14). Statistical analysis assessed by Mann-Whitney U test. Data are presented as mean +s.e.m., ∗p < 0.05.