Nezamabadi, Kasra; Sivalokanathan, Sanjay; Lee, Jiwon; Tanriverdi, Talha; Chen, Meiling; Lu, Daiyin; Abraham, Jadyn; Sardaripour, Neda; Li, Pengyuan; Mousavi, Parvin; Abraham, Maria Roselle. (2025). Explainable artificial intelligence identifies and localizes left ventricular scar in hypertrophic cardiomyopathy using 12-Lead electrocardiogram. Scientific Reports, 15(1), 33918. https://doi.org/10.1038/s41598-025-09282-7
Scarring in the left ventricle (LV) of the heart is a key risk factor for sudden cardiac death and heart failure in people with hypertrophic cardiomyopathy (HCM), a condition where the heart muscle becomes abnormally thick. Because this scarring develops and changes over time, it’s important for doctors to monitor it regularly. Currently, detecting LV scar requires a specialized MRI technique called late gadolinium enhancement, which is accurate but expensive and can’t always be used for patients with implanted heart devices due to image artifacts.
To overcome these challenges, we developed XplainScar, an explainable machine learning model that can detect LV scar using only data from a standard 12-lead electrocardiogram (ECG) — a simple, low-cost test commonly used in clinics. We trained and validated XplainScar using data from 748 patients with HCM collected at two major medical centers (500 from Johns Hopkins Hospital for training and 248 from UCSF for validation). The model uses advanced unsupervised and self-supervised learning techniques to both identify the presence of LV scar and highlight the specific ECG features linked to it.
XplainScar can quickly process ECG data — analyzing 10 patients in under a minute — and showed strong accuracy in testing, with an F1-score of 89%, sensitivity of 90%, specificity of 78%, and precision of 88%. These results suggest that XplainScar provides a fast, affordable, and transparent alternative to MRI for detecting and monitoring LV scar in HCM patients. By improving accessibility and reducing costs, this tool could support better long-term care for individuals with heart disease. The model and code are publicly available at https://github.com/KasraNezamabadi/XplainScar.
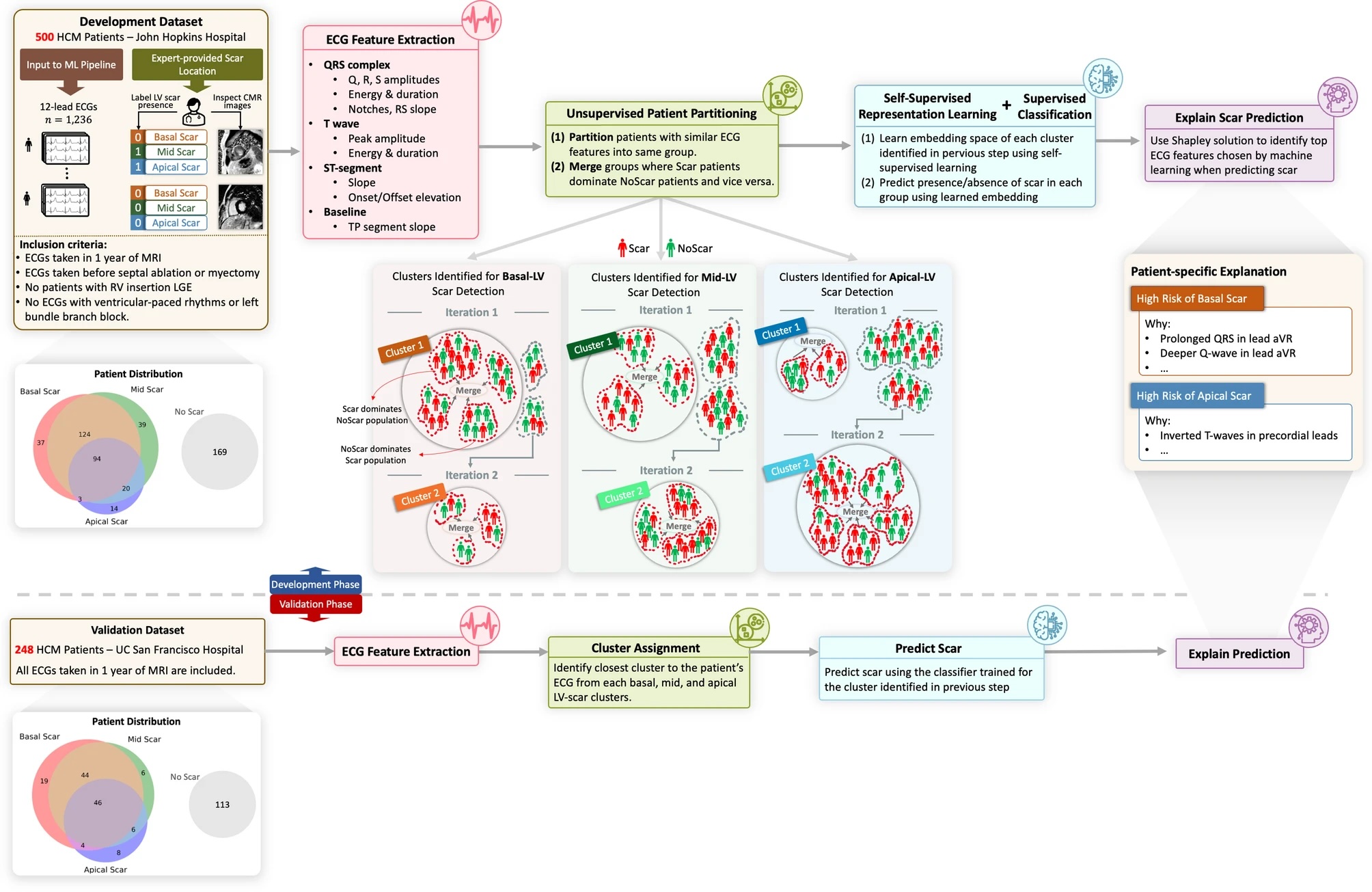
Fig 1
Overview of XplainScar. This method was developed using the JH-HCM dataset (n = 500), and validated using the UCSF-HCM dataset (n = 248), after excluding HCM patients with LGE at RV insertion sites. In each patient, the LV was divided into basal, mid, apical regions for scar (LV-LGE) detection. P, Q, R, S, T waves in 12-lead ECGs were identified using a segmentation method developedfor HCM ECGs. ECG features such as duration, amplitude, slope, energy of QRS complexe and T waves, as well as ST, TP segments were extracted from each lead, and adjusted for LV mass index, age, sex, using multiple linear regression. Subsequently, patients were partitioned into groups based on similarity of their ECG features using unsupervised clustering. In each group, a self-supervised neural network followed by a fully connected neural net predicted presence of LV scar. The Shapley value approach was used to identify the top ECG features that participated in LV scar prediction in the basal, mid and apical LV in each HCM patient.