Zhang, Rendong, Chiron, Sophie, Tyree, Regina N., Carson, Kate S., Raber, Larry W., Ramadass, Karthik, Gao, Chenyu, Kim, Michael E., Zuo, Lianrui, & Li, Yike. (2025). Enhancing clinical data management through barcode integration and research electronic data capture: Scalable and adaptable implementation study. JMIR Formative Research, 9, e70016. https://doi.org/10.2196/70016
Managing data accurately and efficiently is critical in clinical studies, where researchers need to securely track, store, and analyze large numbers of patient samples. Traditional systems often struggle with problems like barcodes not being recognized correctly, lack of detailed information, and slow performance when handling heavy workloads.
In this study, we set out to improve clinical data management by making barcodes more reliable, adding more detailed information to the data, and increasing the speed of the system. To do this, we tested different barcode technologies under difficult conditions, such as blurred images, and found that DataMatrix barcodes were the most resilient. We also created a dynamic organ-specific archive within the REDCap database, which allowed us to more precisely track where each sample came from, adding valuable detail to the data. In addition, we used Docker to package the informatics software so it could be easily deployed and remain consistent across different studies. Finally, we developed a local cache system that greatly reduced the time needed to interact with REDCap when working with large datasets, cutting processing times by nearly eightfold.
Overall, these improvements make clinical data systems more robust, faster, and more precise. They provide a stronger foundation for managing complex data in real-world research, ensuring that studies—such as those focused on gastrointestinal diseases—can be conducted more efficiently and accurately.
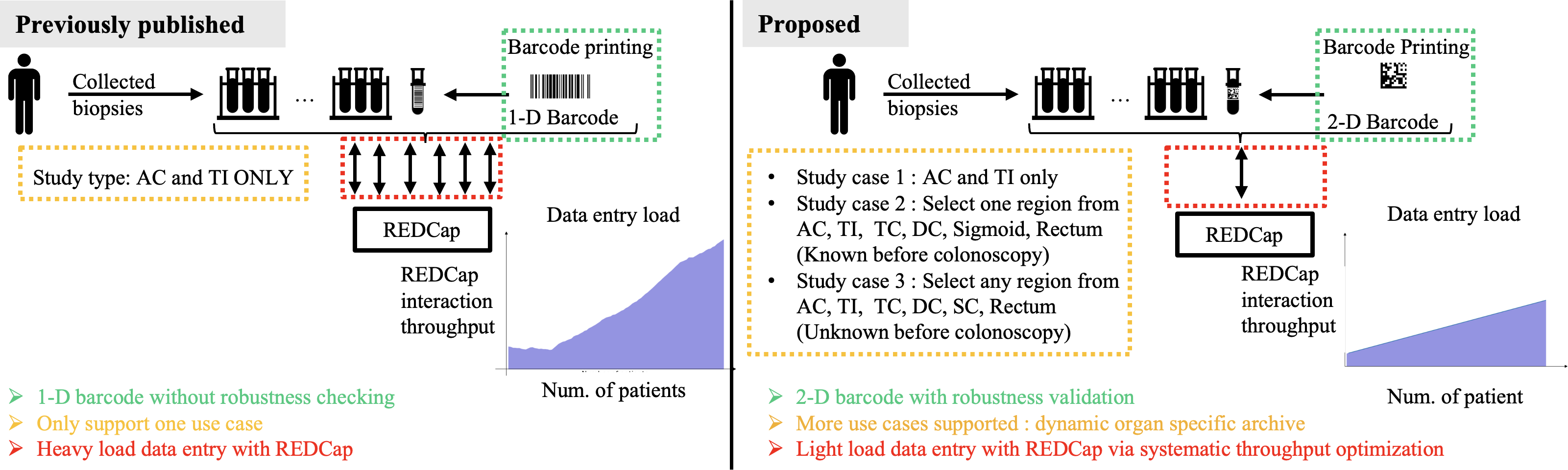
Figure 1. Comparison of the published and proposed barcode systems for collected biopsies. The published system uses 1-D barcodes, supports only one use case (ascending colon [AC] and terminal ileum [TI]), and involves a heavy data entry load with REDCap without robustness checking. In contrast, the proposed system uses 2-D barcodes with robustness validation, supports multiple use cases, including dynamic organ-specific archives, and optimizes data entry with a lighter load on REDCap. For example, Study 1 covered that the published system collects samples only from the AC and TI. In contrast, the newly involved Study 2 involves several cases where each case requires sample collection from a specific region, such as the AC, TI, transverse colon (TC), descending colon (DC), sigmoid colon (SC), or rectum. Study 3 necessitates collecting samples from multiple regions. AC: ascending colon; DC: descending colon; Num.: number; REDCap: Research Electronic Data Capture; TC: transverse colon; TI: terminal ileum; SC: sigmoid colon.