Woodhouse, Palina; Jackson, Laurel; Kammer, Michael N.; Godfrey, Caroline M.; Antic, Sanja; Zou, Yong; Meyers, Patrick; Gawel, Susan H.; Maldonado, Fabien; Grogan, Eric L.; Davis, Gerard J.; Deppen, Stephen A. “Optimizing Biomarker Models for Biologically Heterogeneous Cancers: A Nested Model Approach for Lung Cancer.” Cancer Epidemiology, Biomarkers & Prevention 34, no. 5 (2025): 788–794. https://doi.org/10.1158/1055-9965.EPI-24-0523.
Lung cancer comes in different forms, each with its own unique biology. This makes it hard to create reliable blood tests (called biomarkers) that can detect cancer early. Traditional methods for building these tests often don’t work well across all the different subtypes of lung cancer. This study tested a new approach, called a “nested biomarker model,” to see if it could better handle this complexity and improve early detection.
The study looked at 337 patients from two medical centers. Blood samples were collected and analyzed, and the researchers used advanced statistical methods to create the nested model. This model was designed to recognize the differences among various lung cancer subtypes and was compared to more traditional models.
The patients had a mix of cancerous and non-cancerous lung nodules, covering a range of lung cancer types. The new nested model performed similarly to well-known models like the one used at the Mayo Clinic. It was especially good at identifying small cell lung cancer, one of the more aggressive subtypes.
The study shows that the variety of lung cancer types makes it difficult to create a one-size-fits-all blood test. However, the nested model offers a promising new way to improve early cancer detection by taking this variety into account. More research with larger groups of patients is needed to confirm these results, but this approach could help build better tests for detecting different types of cancer early.
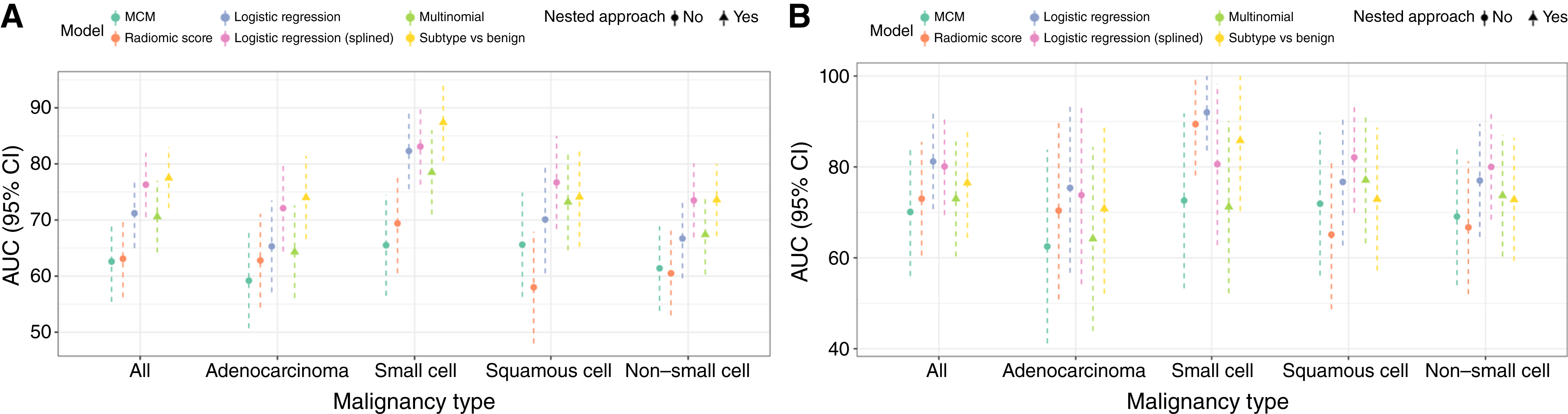
Figure 1.
A, AUCs of evaluated models in training data. AUCs with 95% CIs for all models evaluated in the training dataset, reported for all malignancy types and each subtype, are shown. B, AUCs of evaluated models in testing data. AUCs with 95% CIs for all models evaluated in the testing dataset, reported for all malignancy types and each subtype, are shown.