Gomes, S. F., Jr.; Velasco, K.; Cunha, S.; Santos, J.; Aboelkheir, M. G.; Sumini, M.; Thiré, R.; Duarte, P. C., Jr.; Andrade, A. J. P.; Díaz-Martín, R. D.; Clebis, V. H.; Bhansali, S.; Pal, K.; Maranhão, F. “Leveraging Large Language Models for Accelerated Learning and Innovation in Biogenic Tissue-Engineered Vascular Grafts.” Journal of Drug Delivery Science and Technology 108 (2025): 106935. https://doi.org/10.1016/j.jddst.2025.106935.
This study uses Large Language Models (LLMs)—a type of advanced artificial intelligence—to help speed up progress in developing tissue-engineered vascular grafts (TEVGs), which are lab-made blood vessels used to repair or replace damaged ones. TEVGs face several major challenges, including making sure they’re safe for the body (biocompatibility), allowing living cells to grow and integrate properly, and improving how the grafts are designed (scaffold optimization).
By using LLMs to review and analyze a large amount of scientific literature, the researchers were able to spot important trends and breakthroughs in materials and techniques used to build these grafts. This includes newer fabrication methods like hybrid 3D printing, electrospinning, and melt electrowriting, which help improve how blood flows through the grafts (hemodynamics) and make them mechanically stronger and more natural.
One promising material combination the study highlights is polycaprolactone (PCL) mixed with collagen. These scaffolds support healthy cell growth and tissue remodeling, closely mimicking real blood vessels. The approach also tackles ongoing problems like blood clotting (thrombosis), stiffness mismatches between the graft and the body’s vessels, and poor performance of small-diameter grafts.
By bringing AI into the research process, this study presents a new way to drive innovation in TEVGs and move closer to making them more effective and widely used in clinical settings.
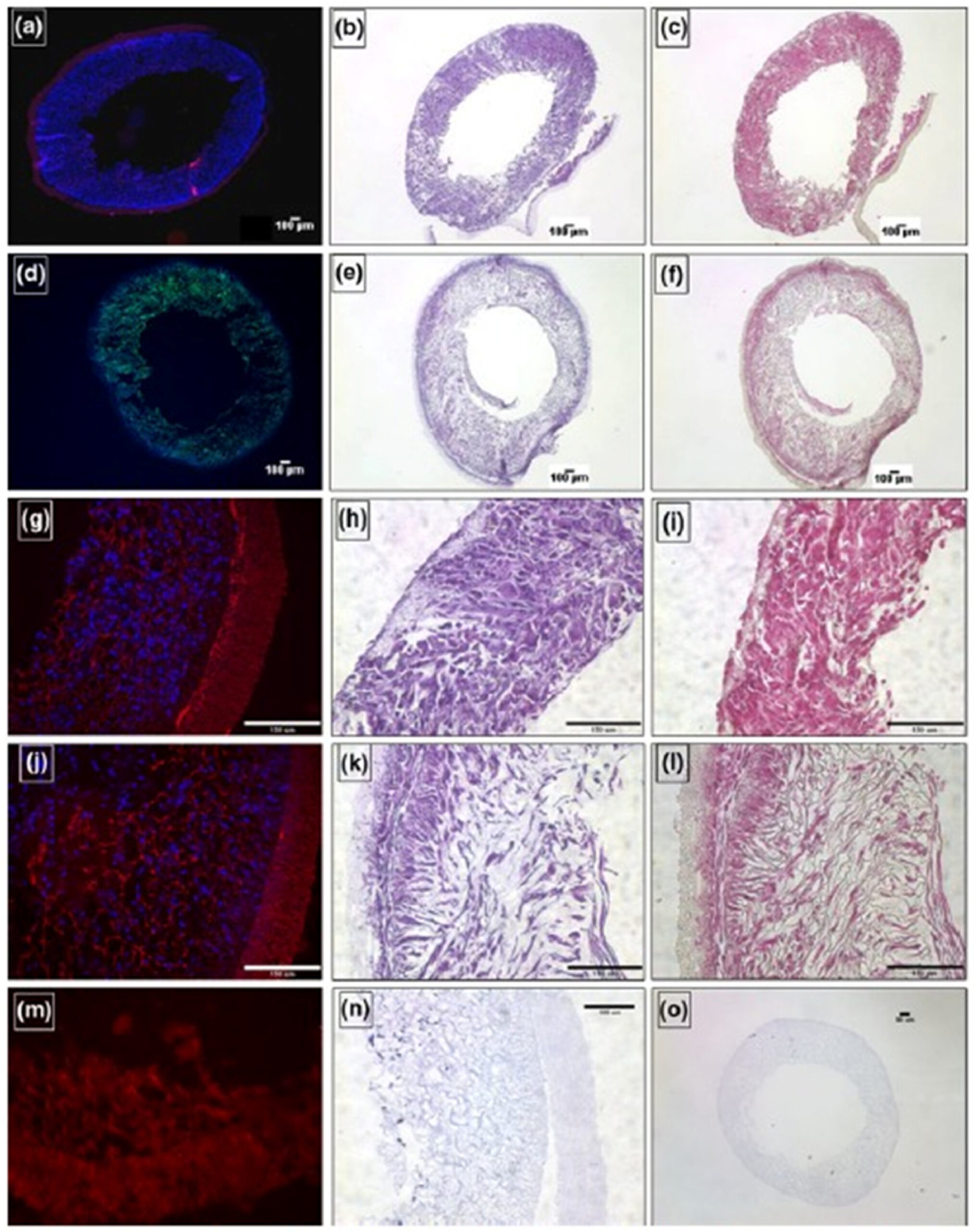
Fig. 1. In vitro characterization of the rSMC-seeded ES-TIPS PEUU constructs (a-c and g-i) immediately after seeding and (d-f and j-i) after dynamic culture for 2 days in a spinner flask. (a, g and j). Nuclear staining (red = autofluorescence of scaffold, blue = nuclei); (b, e, h, k,n and o) H&E staining; (c, f, i and l). Masson’s trichrome staining; (d) F-actin staining (green = F-actin, blue = nuclei). (m–o) unseeded or poorly-seeded ES-TIPS scaffolds. Reprinted with permission from Copyright: © 2017 Ulf Bertram et al. This is an open-access article distributed under the Creative Commons Attribution 4.0 License. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)