Newlin, Nancy; Schilling, Kurt; Koudoro, Serge; Chandio, Bramsh Qamar; Kanakaraj, Praitayini; Moyer, Daniel; Kelly, Claire E.; Genc, Sila; Yang, Joseph Yuan-Mou; Wu, Ye; Adluru, Nagesh; Nath, Vishwesh; Pathak, Sudhir; Schneider, Walter; Gade, Anurag; Consagra, William; Rathi, Yogesh; Hendriks, Tom; Vilanova, Anna; Chamberland, Maxime; Pieciak, Tomasz; Ciupek, Dominika; Vega, Antonio Tristán; Aja-Fernández, Santiago; Malawski, Maciej; Ouedraogo, Gani; Machnio, Julia; Thompson, Paul M.; Jahanshad, Neda; Garyfallidis, Eleftherios; Landman, Bennett. “Introducing QuantConn: Overcoming Challenging Diffusion Acquisitions with Harmonization.” Lecture Notes in Computer Science 15171 LNCS (2025): 164–174. https://doi.org/10.1007/978-3-031-86920-4_15.
Changes in the brain’s white matter are becoming more recognized as important in understanding neurological diseases and how they progress. A special type of brain scan called diffusion-weighted MRI (DW-MRI) is often used in large international studies to look at the structure and connections in white matter. However, it’s hard to compare results from different places because the scans are often done using different machines or methods. This lack of consistency makes it difficult to get reliable data.
To solve this, researchers are working on ways to “harmonize” or standardize how DW-MRI data is processed. This would help ensure that results from different research sites can be trusted and compared. The goal is to get accurate measurements of white matter properties, including:
(1) microstructure details from specific white matter bundles,
(2) the shape and size of those bundles, and
(3) how different parts of the brain are connected (connectivity).
In the MICCAI CDMRI 2023 QuantConn challenge, researchers were given brain scan data from the same people, collected using two slightly different scan methods on the same machine. Their task was to process the data in a way that reduces the differences caused by the scanning method—while keeping the real biological differences between individuals.
This paper explains how the challenge was set up, presents initial results before the data was harmonized, and talks about what researchers can learn from participating in this challenge.
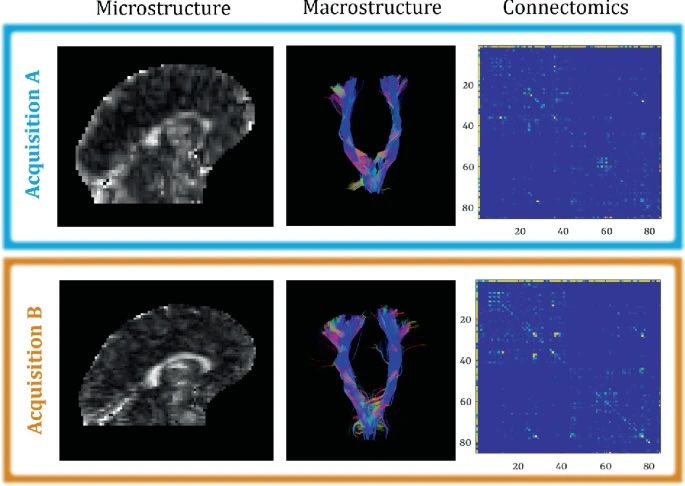
Fig. 1.
Acquisition differences affect microstructure measurements (FA map, left), macrostructure reconstructions (corticospinal tract shape, middle), and connectomics (whole brain connectome, right).