Yang, Yuechen; Wang, Yu; Yao, Tianyuan; Deng, Ruining; Yin, Mengmeng; Zhao, Shilin; Yang, Haichun; Huo, Yuankai. “PySpatial: A High-Speed Whole Slide Image Pathomics Toolkit.” IS and T International Symposium on Electronic Imaging Science and Technology 37, no. 12 (2025): HPCI-177. https://doi.org/10.2352/EI.2025.37.12.HPCI-177.
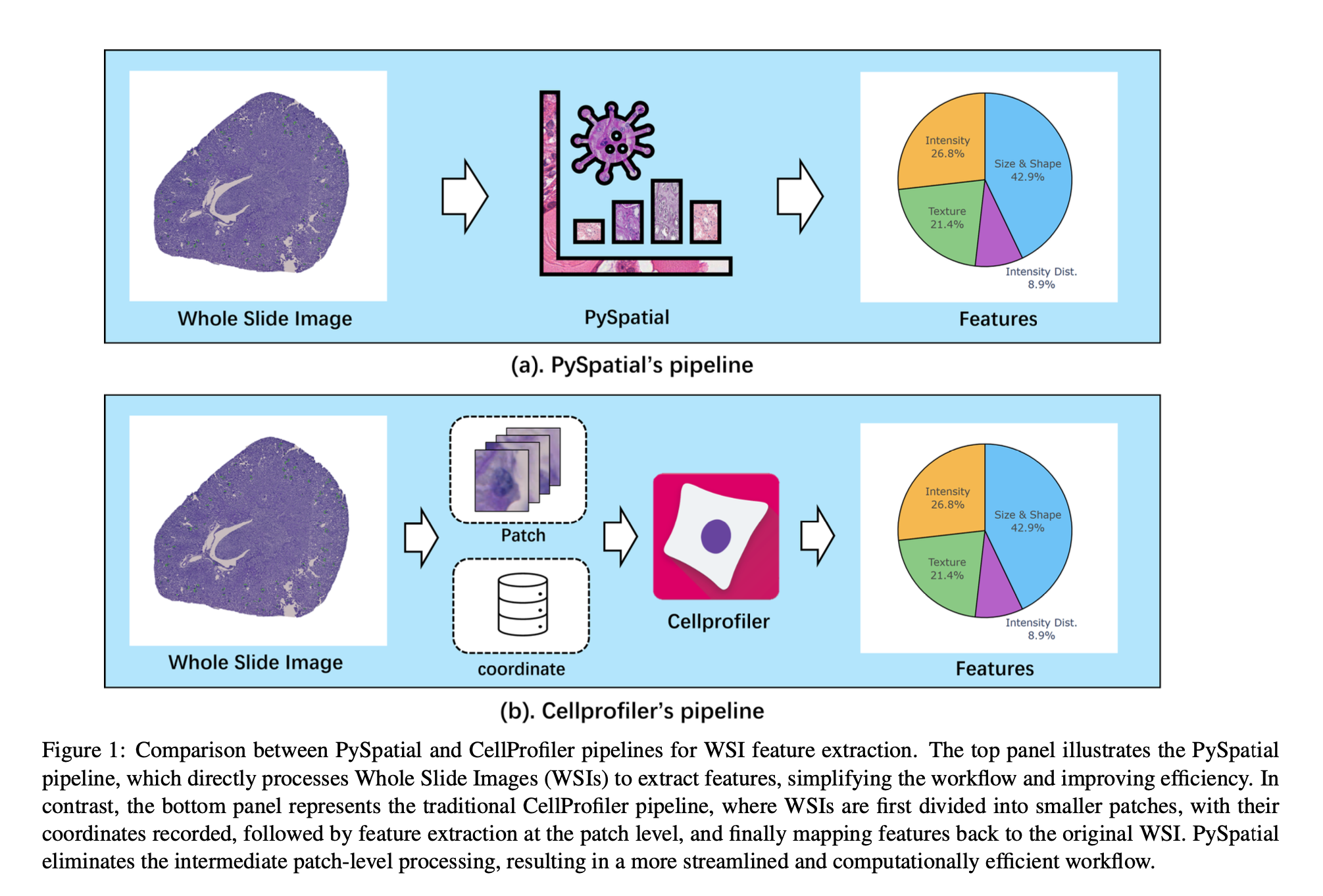
Analyzing Whole Slide Images (WSIs) is an important part of modern digital pathology because it allows researchers to extract many features from tissue samples. However, traditional methods using tools like CellProfiler can be slow and involve several steps: dividing the WSI into smaller patches, extracting features from each patch, and then combining those features back into the full image.
To make this process faster and simpler, we developed PySpatial, a high-speed toolkit designed specifically for analyzing WSIs. PySpatial improves the usual process by working directly on selected regions of the image, which reduces unnecessary steps. It uses special techniques like rtree-based spatial indexing and matrix-based computation to quickly find and process these regions.
We tested PySpatial on two datasets—one involving Perivascular Epithelioid Cell (PEC) tumors and another from the Kidney Precision Medicine Project (KPMP)—and found major improvements in performance. For small and scattered features in the PEC data, PySpatial was almost 10 times faster than CellProfiler. For larger structures like glomeruli and arteries in the KPMP data, it was twice as fast.
These results show that PySpatial can speed up large-scale WSI analysis while keeping accuracy high, making it a useful tool for advancing digital pathology.