Green, Emily H.; Kotrannavar, Subhag R.; Rutherford, Megan E.; Lunnemann, Hannah M.; Kaur, Harsimran; Heiser, Cody N.; Ding, Hua; Simmons, Alan J.; Liu, Xiao; Lacy, D. Borden; Washington, M. Kay; Shrubsole, Martha J.; Liu, Qi; Lau, Ken S.; Sears, Cynthia L.; Coffey, Robert J.; Drewes, Julia L.; Markham, Nicholas O. “Multiomic spatial atlas shows deleted in malignant brain tumors 1 (DMBT1) glycoprotein is lost in colonic dysplasia.” Journal of Pathology 266, no. 1 (2025): 51-65. https://doi.org/10.1002/path.6406.
Colorectal cancer (CRC) causes over 900,000 deaths each year around the world. New research suggests that certain harmful bacteria in the colon might help promote or even cause CRC. In earlier studies, we found that a bacteria called Clostridioides difficile (C. difficile), which is commonly found in the gut of people with CRC, can speed up the development of tumors in mice. To better understand how these bacteria interact with the body during tumor growth, we used advanced techniques to study the genes and proteins in the colon. We discovered that a gene called DMBT1 behaves very differently in inflamed versus tumor-affected areas of the colon. When we looked at the gene expression in healthy colon cells exposed to C. difficile, we found that the bacteria caused DMBT1 to be more active. However, in areas where tumors were developing, this gene was almost completely turned off. We confirmed these findings with various experiments and found that DMBT1 was also turned off in tumor samples from both mice and humans. Further tests showed that a cell signaling process called WNT might be responsible for this change in the gene’s activity. Overall, our research suggests that the DMBT1 gene could be a key link between bacteria and the development of colon cancer.
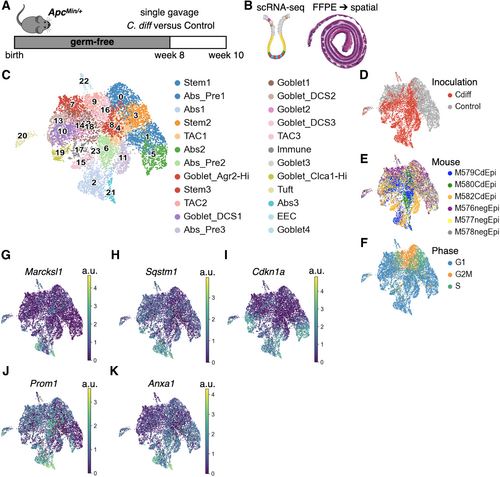
Figure 1
Cell type identification and analysis of scRNA-seq from colonic epithelial cells in C. difficile-colonised ApcMin/+mice demonstrated unique cell states.
(A) Schematic of experimental design: 8-week-old germ-free ApcMin/+ mice were gavaged with C. difficile-containing bacterial consortium or control consortium as previously published [13], and mice were euthanized at 10 weeks. (B) Epithelial crypts were dissociated from the colonic tissue for scRNA-seq; n = 3 mice/group. Spatial transcriptomics was performed on formalin-fixed, paraffin-embedded whole colon; separate experiment with n = 3 mice/group. (C) UMAP plot with cell type assignments for clusters based on canonical mouse gene markers (supplementary material, Figure S2). (D–F) UMAP plots colored by inoculation (C. difficile or control), mouse, or cell cycle phase. (G–K) UMAP plots colored by scaled gene expression (a.u. = arbitrary units) of canonical gene markers for different cell states: fetal reversion (Marcksl1), autophagy (Sqstm1), senescence (Cdkn1), intestinal plasticity (Prom1), and regenerative stemness (Anxa1).