Software
Software our lab has access to |
|
 Adobe Acrobat 8 Professional
Adobe Acrobat 8 Professional
A useful program for organizing and viewing pdf files. Adobe Acrobat 8 Professional allows you to take multiple pdf files and combing/rearrange them into one complete pdf file. Additionally Acrobat allows you to highlight, edit, and comment within text in a pdf file and save those alterations. These features are unique to the full Adobe Acrobat program and are not offered with the freeware version - Adobe Reader.
|
 AMBER 9.0, 10.0
AMBER 9.0, 10.0
AMBER is a Unix based suite of programs designed for efficient structure
building and molecular dynamics. Our group uses various facets of this
suite such as LEaP, antechamber, and sander for building, minimization and
conformational sampling of structures of empirically studied biomolecular
ions (lipids, nucleotides, natural products). This data is used to
elucidate experimental measurements by calculating the prevailing
conformational arrangements of biomolecular ions in the gas phase.
|
|
 Antechamber
Antechamber
antechamber is used as a preparatory tool for LEaP. It allows the
conversion of various file types, as well as the generation of Amber prep
files using restrained electrostatic potential charge (RESP) fitting for
LEaP. Additionally, Antechamber assigns atom types based on the user
selected force field.
 LEaP
LEaP
LEaP is used to prepare input files for AMBER based molecular dynamics
studies. Biomolecular ions of interest can be built in LEaP. LEaP also
generates topology, parameter, and internal coordinate files for sander.
 sander
sander
sander is used for energy minimization and conformational sampling. The
force field used by Sander during conformational sampling is a set of
equilibrium functions that describe bond, bond angles, electrostatic and
other non-covalent interactions based on classical (Newtonian) physics
derivations. An example set of such functions is shown below:
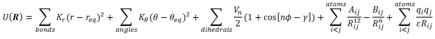
Molecular dynamics based conformational sampling is achieved by heating a
given structure to high temperature, then cooling random high temperature
snapshots very gradually to a low temperature plateau. Cooled structures
can be compared based on their relative energies, conformation and
theoretical collision cross section generated by MOBCAL (see below).
|
 ChemBioOffice 2008
ChemBioOffice 2008
ChemBioOffice is a suite of programs that allow you to
draw, analyze, fragment, etc. molecules. This program suite is available free through Vanderbilt ITS and Chemistry Department. You can download it here after entering your Vanderbilt email address and site license information that you can get from the department office. This suite include ChemBioDraw, ChemBio3D, and a few different ChemFinder programs. The latter of which may only find limited utility in our lab.
|
|
 ChemBioDraw
ChemBioDraw
ChemBioDraw allows for visualization of molecules. There are many ways to construct possible analytes that you may encounter. One way is to use one of the many templates available (seen as a button on the left hand side toolbar) to construct peptides, oligonucleotides, oligosaccharides etc. Similarly you can also press
"Ctrl + Shift + n" and bring up a convert name to structure function that is incredibly useful. Structures that are copied from a ChemDraw workbook and pasted into recent releases of Microsoft Office programs (Word, Excel, Powerpoint) can be edited post-paste by double clicking on the structure. This is useful when having to move, shift, or rearrange structures so that you don't have to return to the root file to modify the molecule. This program primarily renders structures in Fisher style projections and so can not be used for anything beyond primary structural analysis.
 ChemBio3D
ChemBio3D
ChemBio3D as its name implies is more suited for 3-dimensional visualization of molecules. It can accept structures straight from ChemBioDraw either imported or as straight copy-and-paste. This program allows you to import and export structure files as pdb format which can be quite useful when quickly drawing up a structure to export to AMBER or other programs where drawing from scratch is not as easy. It also provides some molecular dynamics capabilities (the effectiveness of which has yet to be determined by our lab).
|
 CorelDraw Graphics Suite X3
CorelDraw Graphics Suite X3
CorelDraw is the go-to program for preparing figures and touching up and modifying graphs for publications. The learning curve is a bit steep for this program, but once you get the hang of it your figures will improve dramatically. When combined with the bundled Corel Photo-Paint X3 there is very little that cannot be done graphically, although Photo-Paint is geared more towards handling and altering photos.
|
 Data Explorer 4.3
Data Explorer 4.3
Data Explorer is the data analysis software that works with the 'Voyager Control Panel' on the Voyager STR instrument. Although it is useful for viewing and analyzing data you may want to export to Origin to allow full customization of plotted spectra. This software has shown perfect compatability with Windows XP, however, when using Vista various portions of installation require you to temporarily turn User Account Control (UAC) off and/or restart the computer multiple times before first use.
|
 Gaussian 03
Gaussian 03
Gaussian is a powerful program for performing quantum mechanical
calculations. In our lab it is used primarily for structure minimization,
frequency and electrostatic potential calculations. Gaussian allows
quantum mechanical treatment of molecules with user defined basis sets.
|
 MassLynx 4.0
MassLynx 4.0
MassLynx 4.0 is suite of programs that our lab uses primarily to view data from the Synapt instrument. There are several programs that can be used when visualizing this type of data. The 'Chromatogram' program opens a spectral trace of the ion current throughout the duration of the sample run. Upon right clicking and dragging over a portion of this trace a 2-D (m/z vs. intensity) graph is generated in 'Spectrum' from the selected data. From here it is possible to analyze and also export this data in various forms. You can also copy the peak list and paste it into a notebook file and then import into programs such as Excel or Origin to allow for more customization of plots. The last program that will be mentioned in this package is DriftScope which is used for visualizing 3-D IM-MS data. Recently DriftScope 2.0 was released which included some additional options such as flipping the axes to allow m/z to be on the abscissa and arrival time distribution to be on the ordinate. It is also now possible to remove the lines from the plot area.
|
 MOBCAL
MOBCAL
MOBCAL is a Fortran program developed in Martin Jarrold’s group at
Northwestern University. Mobcal calculates collision cross section based
on structure of input molecules using either a projection model or
trajectory calculation (based on hard sphere or Lennard Jones potential).
Mobcal is especially recommended for collision cross section calculations
of intermediate and large size molecules. |
 Origin 6.1
Origin 6.1
Origin is a essential tool when modifying spectral traces from various sources and in various formats to create publication worthy graphs and plots. Often the peak lists from programs such as Data Explorer are too large to be handled by the 1,048,576 cells that Excel provides. By saving the data as an ASCII file and using the 'import' function in Origin you can create a plot from the data and alter all aspects of the graph (axis titles, font size, line thickness, graph type, compound graphs, etc.). These graphs can then be exported as many file types (jpg, gif, eps, tif). The software package also includes extensive data analysis tools such as statistics, signal processing, curve fitting, and peak analysis. Similar to Data Explorer this program seems to work fine with Windows XP but Vista may require you to turn User Account Control (UAC) off or restart the computer several times before you can boot it up the first time. |
 SIMION 8.0
SIMION 8.0
SIMION is a program that is used to render potential arrays and simulate the effects of these arrays on preselected groups of ions. We use this software to simulate and optimize preexisting instrumentation in our lab and also to create and test new instrumentation prior to construction.
|
|
|