Michael P. Stone
Professor of Chemistry
Dr. Stone is currently accepting graduate students for Fall 2024 - APPLY TODAY
| Our lab focuses upon chemistry and structural biology of DNA damage, and its role in cancer etiology. We probe chemical rearrangements of initially formed DNA damage, which may alter genotoxic response in cellular DNA. We are particularly interested in the chemistry of agents that alkylate DNA at the N7-dG position; these may re-arrange into chemically reactive apurinic (AP) sites. They also rearrange to formamidopyrmimidine (Fapy-dG) lesions. which exist as a complex mixture of configurational and conformational isomers. We are interested in how DNA repair and replication enzymes process these damage sites in DNA. We utilize a combination NMR spectroscopy and crystallography for structural determination. |
- How do specific DNA adducts alter the three dimensional structure of DNA?
- How does adduction affect the biological processing of DNA during replication and repair?
- How do specific DNA adducts alter the three dimensional structure of primer-template complexes with damage bypass polymerases?
- What is the role of DNA sequence in modulating adduct conformation and biological processing?
Both NMR spectroscopy and X-ray crystallography play major roles in our research program. Our laboratory is affiliated with the Vanderbilt Center in Structural Biology. NMR spectroscopy is used to determine the three dimensional structures and dynamics of site-specifically adducted oligodeoxynucleotides in aqueous solution. Crystallographic approaches allow the structural determination of larger biomolecular complexes involving DNA processing enzymes.
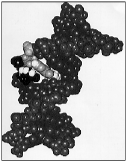
Space-filling model of an oligodeoxynucleo-tide containing a mutagenic lesion, 1,N2 -propanodeoxyguanosine. The model was refined using experimental NOE constraints derived from 1H NMR spectroscopy.
Specializations
| Biophysical Chemistry Chemical Biology Structural Biology Physical Chemistry |
Representative Publications
0000-0002-0922-0216
|
|
|
Bamberger, S.N., Malik, C.K., Johnson-Salyard, T.L., Brown, S.K., Hope, P., Brown, K.L., Li, L., Dempster, R.K., Bowen, R., Voehler, M.W., Rizzo, C.J., & Stone, M.P. "Configurational and Conformational Equilibria of the N6-(2-Deoxy-D-erythropentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylformamidopyrimidine (MeFapy-dG) Lesion in DNA." (2018) Chem. Res. Toxicol. 31, 924-935. PMID: 30169026 PMCID: PMC6352716.
Kellum, A.H., Jr., Qiu, D.Y., Voehler, M.W., Martin, W.J., Gates, K.S., & Stone, M.P. "Structure of a Stable Interstrand DNA Crosslink Involving a beta-N-Glycosyl Linkage Between an N6-dA Amino Group and an Abasic Site." (2021) Biochemistry 60, 41-52. PMID: 33382597 PMCID: PMC7952047
Tomar, R., Minko, I.G., Sharma, P., Lei, L., Kellum, A.H., Jr., Egli, M., Iverson, T.M., McCullough, A.M., Lloyd, R.S., & Stone, M.P "Base Excision Repair of the N-(2-deoxy-D-erythro-pentofuranosyl)-urea Lesion by the hNEIL1 Glycosylase." (2023) Nucleic Acids Res. 51, 3754-3769. PMID: 37014002. PMCID: PMC10164570 |